Gene Information
|
Gene Name
|
CDH8 |
|
Gene ID
|
1006
|
|
Gene Full Name
|
cadherin 8 |
|
Gene Alias
|
Nbla04261 |
|
Transcripts
|
ENSG00000150394
|
|
Virus
|
HBV |
|
Gene Type
|
protein-coding |
|
HPA Location Info
|
Plasma membrane; 
|
|
Membrane Info
|
Predicted intracellular proteins, Predicted membrane proteins |
|
Uniport_ID
|
P55286
|
|
HGNC ID
|
HGNC:1767
|
|
OMIM ID
|
603008 |
|
Summary
|
This gene encodes a type II classical cadherin from the cadherin superfamily, integral membrane proteins that mediate calcium-dependent cell-cell adhesion. Mature cadherin proteins are composed of a large N-terminal extracellular domain, a single membrane-spanning domain, and a small, highly conserved C-terminal cytoplasmic domain. The extracellular domain consists of 5 subdomains, each containing a cadherin motif, and appears to determine the specificity of the protein's homophilic cell adhesion activity. Type II (atypical) cadherins are defined based on their lack of a HAV cell adhesion recognition sequence specific to type I cadherins. This particular cadherin is expressed in brain and is putatively involved in synaptic adhesion, axon outgrowth and guidance. [provided by RefSeq, Jul 2008] |
Target gene [CDH8] related to VISs 
Integration Table: if previous studies reported that target gene was altered by virus integration events, the overlap between VISs in this literature and Cistrome factors was listed in this section
| DVID |
Chromosome |
HM |
TFBS |
CA |
Sum of Overlapped Records |
Detail |
| 1003528 |
chr16 |
54 |
67 |
5 |
126 |
View |
| 1010521 |
chr16 |
9 |
2 |
1 |
12 |
View |
| 1015617 |
chr16 |
2 |
0 |
0 |
2 |
View |
| 1019608 |
chr16 |
0 |
0 |
0 |
0 |
View |
| 1022720 |
chr16 |
54 |
67 |
5 |
126 |
View |
| 1041167 |
chr16 |
0 |
0 |
0 |
0 |
View |
Target gene [CDH8] related to Omics data
| Data ID |
Experiment type |
Sample number |
Platform |
|
GSE236281
|
RNA-seq |
12 |
Illumina MiSeq (Homo sapiens) |
|
C GSE35465
|
Chip-seq;RNA-seq |
6 |
Illumina HiSeq 2000 (Homo sapiens) |
|
C GSE68402
|
Chip-seq |
26 |
Illumina MiSeq (Homo sapiens);Illumina HiSeq 2500 (Homo sapiens) |
|
TCGA_LIHC_HBV
|
DNA methylation sequencing;RNA-seq |
97 |
TCGA |
|
C GSE270130
|
Chip-seq |
27 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE224901
|
RNA-seq |
21 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
E C GSE100400
|
Chip-seq;RNA-seq;4C_cccDNA |
31 |
Illumina NextSeq 500 (Homo sapiens);Illumina NextSeq 500 (Mus musculus) |
|
GSE173897
|
RNA-seq |
95 |
Illumina HiSeq 4000 (Homo sapiens) |
|
GSE262515
|
RNA-seq |
21 |
Illumina HiSeq 2500 (Homo sapiens);Illumina HiSeq 2500 (Mus musculus) |
|
GSE110345
|
RNA-seq |
4 |
Illumina HiSeq 2500 (Homo sapiens) |
|
C GSE131257
|
ATAC-seq;RNA-seq |
19 |
Illumina HiSeq 2500 (Homo sapiens) |
|
GSE94660
|
RNA-seq |
42 |
Illumina HiSeq 2500 (Homo sapiens) |
When the query gene is differentially changed in the dataset, a volcano/bar plot will be displayed.
> Dataset: GSE100400 - CDH8 expression across samples
|
Volcano Plot
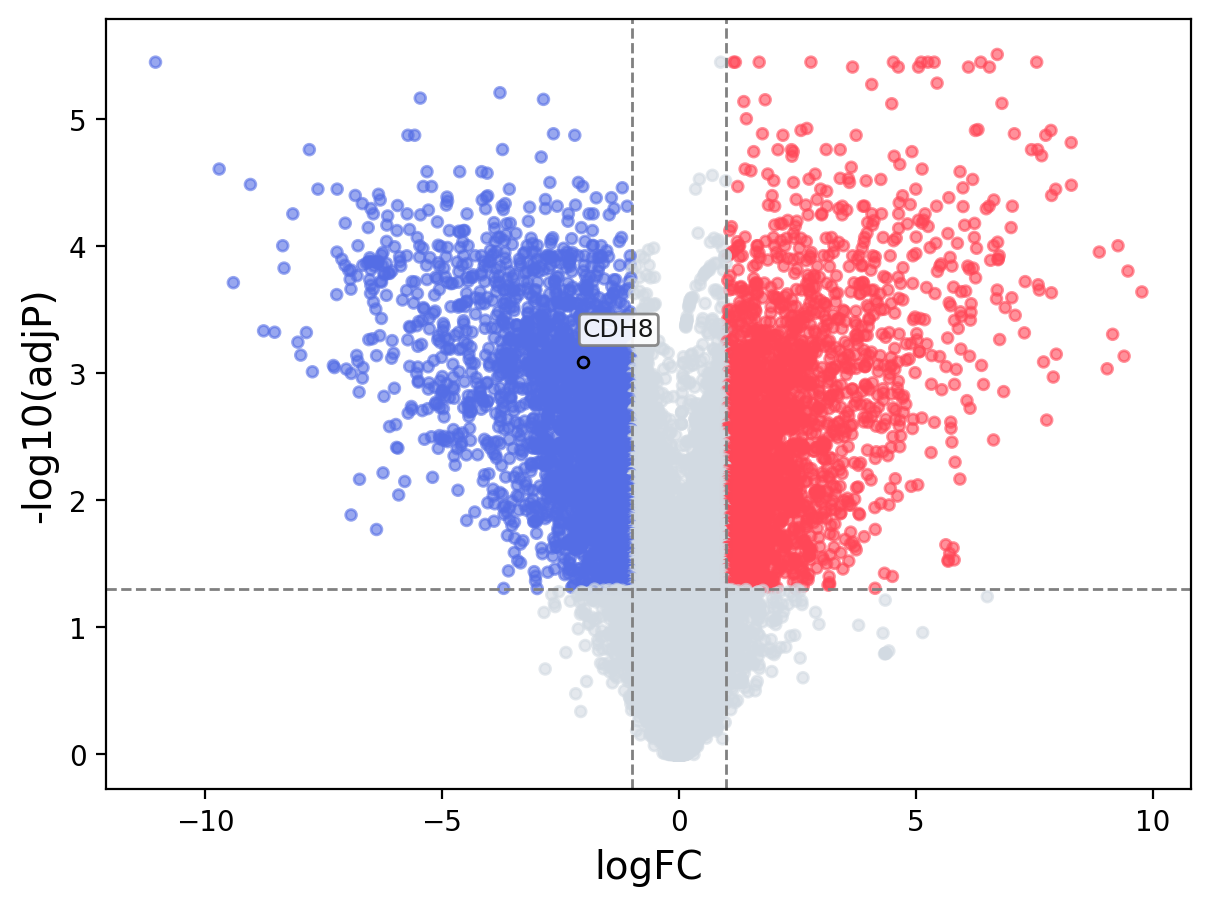
|
Bar Plot
|
|
|
 HBV Target gene Detail Information
HBV Target gene Detail Information