Gene Information
|
Gene Name
|
CFHR5 |
|
Gene ID
|
81494
|
|
Gene Full Name
|
complement factor H related 5 |
|
Gene Alias
|
CFHL5|CFHR5D|FHR-5|FHR5 |
|
Transcripts
|
ENSG00000134389
|
|
Virus
|
HBV |
|
Gene Type
|
protein-coding |
|
HPA Location Info
|
Secreted to blood; 
|
|
Membrane Info
|
Disease related genes, Human disease related genes, Plasma proteins, Predicted intracellular proteins, Predicted secreted proteins |
|
Uniport_ID
|
Q9BXR6
|
|
HGNC ID
|
HGNC:24668
|
|
OMIM ID
|
608593 |
|
Summary
|
This gene is a member of a small complement factor H (CFH) gene cluster on chromosome 1. Each member of this gene family contains multiple short consensus repeats (SCRs) typical of regulators of complement activation. The protein encoded by this gene has nine SCRs with the first two repeats having heparin binding properties, a region within repeats 5-7 having heparin binding and C reactive protein binding properties, and the C-terminal repeats being similar to a complement component 3 b (C3b) binding domain. This protein co-localizes with C3, binds C3b in a dose-dependent manner, and is recruited to tissues damaged by C-reactive protein. Allelic variations in this gene have been associated, but not causally linked, with two different forms of kidney disease: membranoproliferative glomerulonephritis type II (MPGNII) and hemolytic uraemic syndrome (HUS). [provided by RefSeq, Jan 2010] |
Target gene [CFHR5] related to VISs 
Integration Table: if previous studies reported that target gene was altered by virus integration events, the overlap between VISs in this literature and Cistrome factors was listed in this section
| DVID |
Chromosome |
HM |
TFBS |
CA |
Sum of Overlapped Records |
Detail |
| 1041754 |
chr1 |
42 |
8 |
2 |
52 |
View |
Target gene [CFHR5] related to Omics data
| Data ID |
Experiment type |
Sample number |
Platform |
|
GSE236281
|
RNA-seq |
12 |
Illumina MiSeq (Homo sapiens) |
|
C GSE35465
|
Chip-seq;RNA-seq |
6 |
Illumina HiSeq 2000 (Homo sapiens) |
|
C GSE68402
|
Chip-seq |
26 |
Illumina MiSeq (Homo sapiens);Illumina HiSeq 2500 (Homo sapiens) |
|
TCGA_LIHC_HBV
|
DNA methylation sequencing;RNA-seq |
97 |
TCGA |
|
C GSE270130
|
Chip-seq |
27 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE224901
|
RNA-seq |
21 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE100400
|
Chip-seq;RNA-seq;4C_cccDNA |
31 |
Illumina NextSeq 500 (Homo sapiens);Illumina NextSeq 500 (Mus musculus) |
|
GSE173897
|
RNA-seq |
95 |
Illumina HiSeq 4000 (Homo sapiens) |
|
GSE262515
|
RNA-seq |
21 |
Illumina HiSeq 2500 (Homo sapiens);Illumina HiSeq 2500 (Mus musculus) |
|
GSE110345
|
RNA-seq |
4 |
Illumina HiSeq 2500 (Homo sapiens) |
|
C GSE131257
|
ATAC-seq;RNA-seq |
19 |
Illumina HiSeq 2500 (Homo sapiens) |
|
GSE94660
|
RNA-seq |
42 |
Illumina HiSeq 2500 (Homo sapiens) |
When the gene can detect a peak in the dataset, a peak plot will be displayed.
> Dataset: GSE35465 - CFHR5 peak across samples
|
Peak Plot
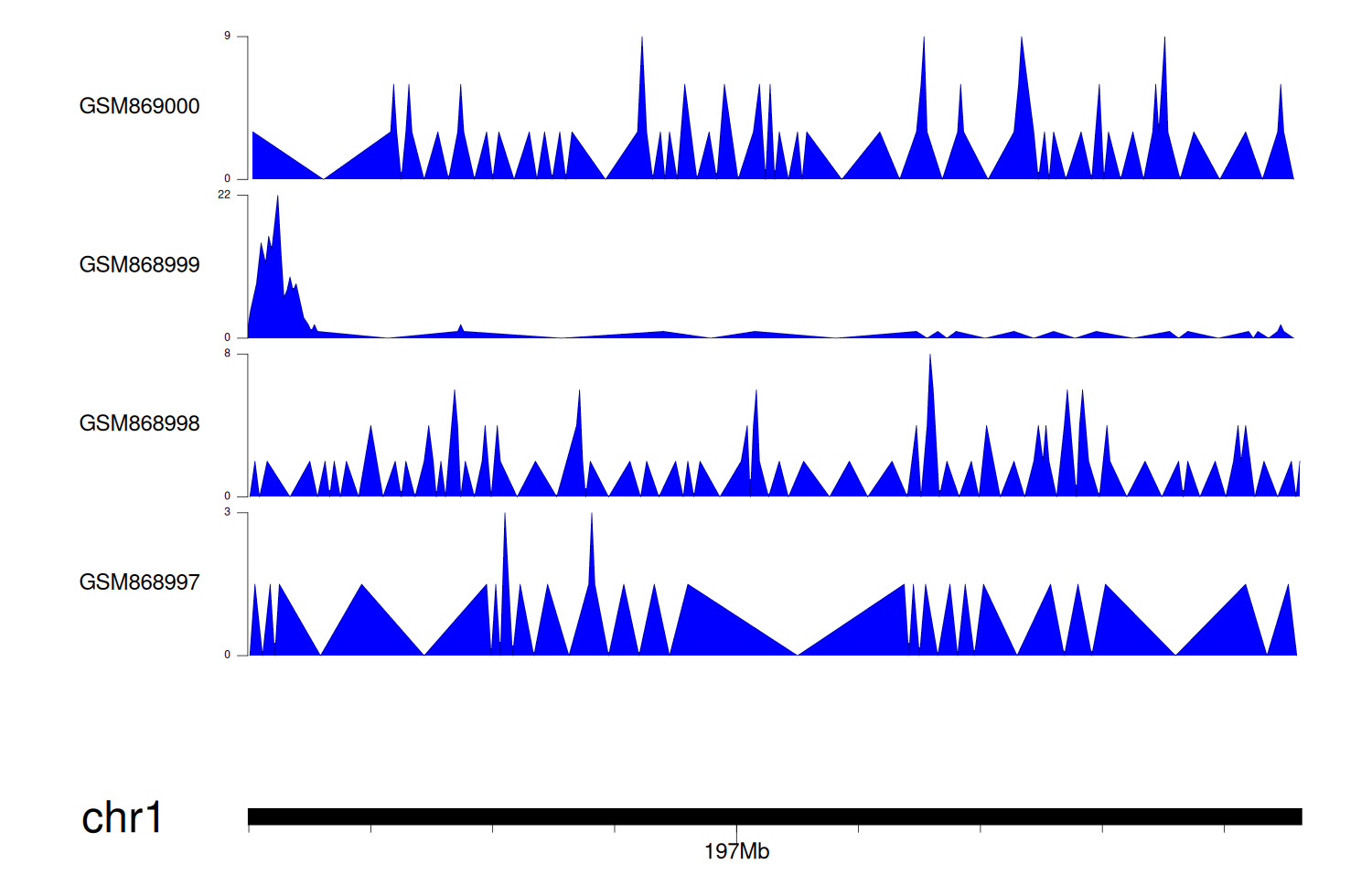
|
> Dataset: GSE68402 - CFHR5 peak across samples
|
Peak Plot
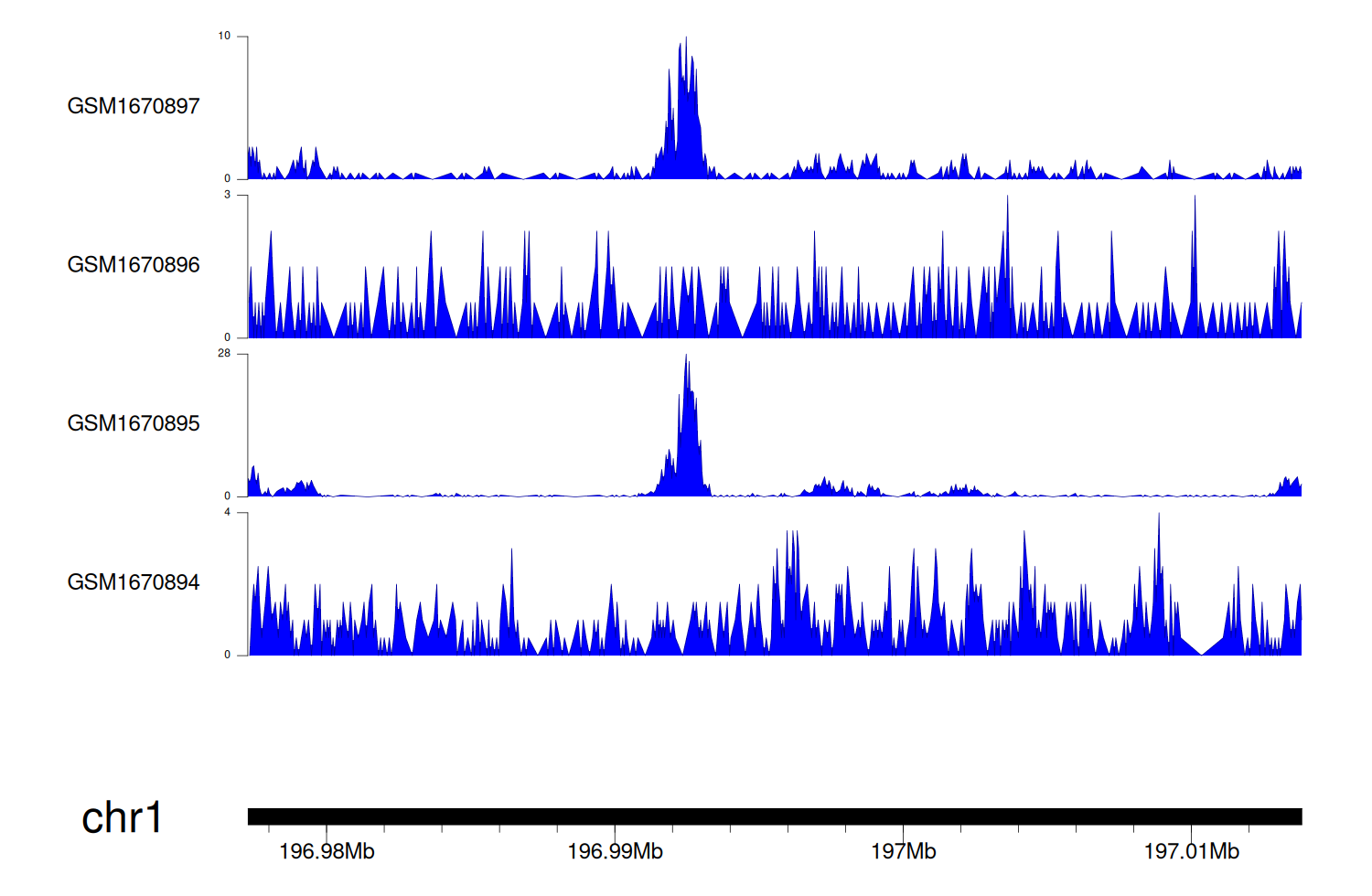
|
> Dataset: GSE270130 - CFHR5 peak across samples
|
Peak Plot
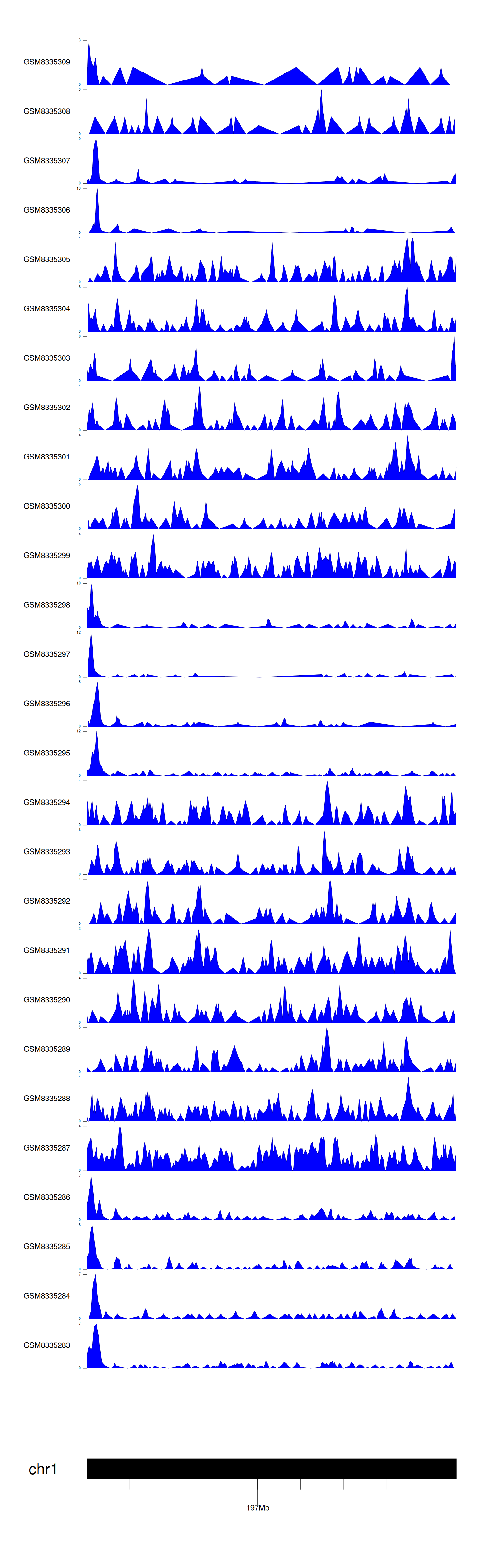
|
> Dataset: GSE131257 - CFHR5 peak across samples
|
Peak Plot
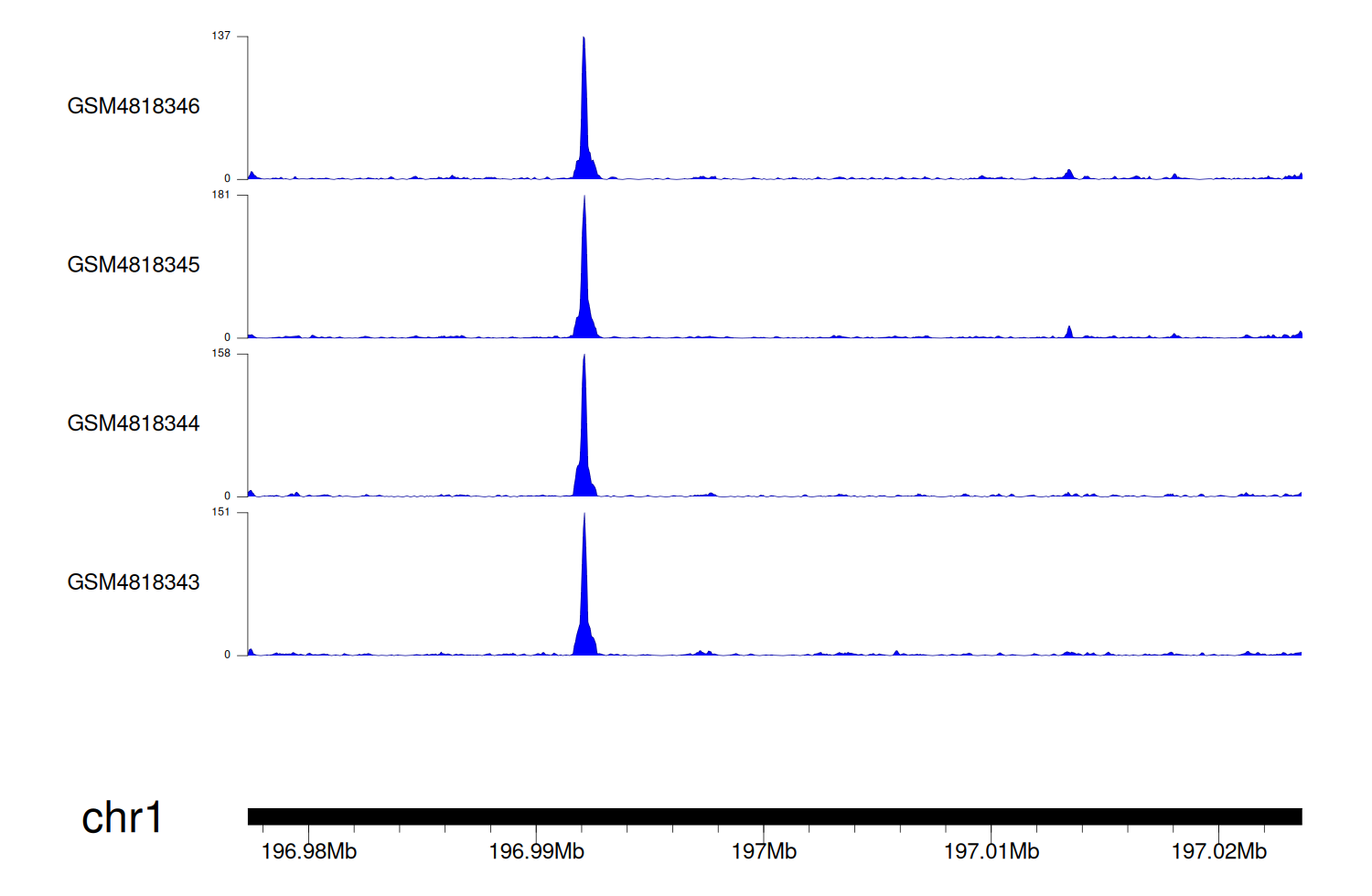
|
|
|
 HBV Target gene Detail Information
HBV Target gene Detail Information