Gene Information
|
Gene Name
|
DDX3X |
|
Gene ID
|
1654
|
|
Gene Full Name
|
DEAD-box helicase 3 X-linked |
|
Gene Alias
|
CAP-Rf|DBX|DDX14|DDX3|HLP2|MRX102|MRXSSB |
|
Transcripts
|
ENSG00000215301
|
|
Virus
|
HPV |
|
Gene Type
|
protein-coding |
|
HPA Location Info
|
Cytosol; 
|
|
Membrane Info
|
Cancer-related genes, Disease related genes, Enzymes, Human disease related genes, Plasma proteins, Potential drug targets, Predicted intracellular proteins, Predicted membrane proteins, Transporters |
|
Uniport_ID
|
O00571
|
|
HGNC ID
|
HGNC:2745
|
|
OMIM ID
|
300160 |
|
Summary
|
The protein encoded by this gene is a member of the large DEAD-box protein family, that is defined by the presence of the conserved Asp-Glu-Ala-Asp (DEAD) motif, and has ATP-dependent RNA helicase activity. This protein has been reported to display a high level of RNA-independent ATPase activity, and unlike most DEAD-box helicases, the ATPase activity is thought to be stimulated by both RNA and DNA. This protein has multiple conserved domains and is thought to play roles in both the nucleus and cytoplasm. Nuclear roles include transcriptional regulation, mRNP assembly, pre-mRNA splicing, and mRNA export. In the cytoplasm, this protein is thought to be involved in translation, cellular signaling, and viral replication. Misregulation of this gene has been implicated in tumorigenesis. This gene has a paralog located in the nonrecombining region of the Y chromosome. Pseudogenes sharing similarity to both this gene and the DDX3Y paralog are found on chromosome 4 and the X chromosome. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Oct 2014] |
Target gene [DDX3X] related to VISs 
Integration Table: if previous studies reported that target gene was altered by virus integration events, the overlap between VISs in this literature and Cistrome factors was listed in this section
| DVID |
Chromosome |
HM |
TFBS |
CA |
Sum of Overlapped Records |
Detail |
| 5008328 |
chrX |
13 |
4 |
0 |
17 |
View |
| 5013919 |
chrX |
100 |
48 |
0 |
148 |
View |
| 5013920 |
chrX |
106 |
87 |
9 |
202 |
View |
| 5016556 |
chrX |
25 |
14 |
0 |
39 |
View |
Target gene [DDX3X] related to Omics data
| Data ID |
Experiment type |
Sample number |
Platform |
|
C GSE183048
|
Chip-seq |
24 |
Illumina HiSeq 4000 (Homo sapiens) |
|
GSE226620
|
scRNA-seq |
30 |
HiSeq X Ten (Homo sapiens) |
|
GSE40774
|
Expression array |
134 |
Agilent-026652 Whole Human Genome Microarray 4x44K v2 (Probe Name version) |
|
GSE139324
|
scRNA-seq |
63 |
Illumina NextSeq 500 (Homo sapiens) |
|
GSE65858
|
Expression array |
270 |
Illumina HumanHT-12 V4.0 expression beadchip |
|
GSE181805
|
Expression array |
25 |
[HTA-2_0] Affymetrix Human Transcriptome Array 2.0 [transcript (gene) version] |
|
GSE196215
|
RNA-seq |
8 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE164690
|
scRNA-seq |
51 |
Illumina NextSeq 500 (Homo sapiens) |
|
GSE138080
|
Expression array |
35 |
Agilent-014850 Whole Human Genome Microarray 4x44K G4112F (Feature Number version) |
|
GSE140662
|
Expression array |
8 |
[HTA-2_0] Affymetrix Human Transcriptome Array 2.0 [transcript (gene) version] |
|
GSE55542
|
Expression array |
36 |
Agilent-039494 SurePrint G3 Human GE v2 8x60K Microarray 039381 (Probe Name version) |
|
C GSE143026
|
ATAC-seq;Chip-seq;RNA-seq |
30 |
Illumina HiSeq 2500 (Homo sapiens) |
|
TCGA_CESC
|
DNA methylation sequencing;RNA-seq |
288 |
TCGA |
|
GSE51993
|
Expression array |
48 |
Illumina Human v2 MicroRNA expression beadchip;Illumina HumanHT-12 V4.0 expression beadchip |
|
S GSE197461
|
scRNA-seq |
16 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE55550
|
Expression array |
155 |
Agilent-039494 SurePrint G3 Human GE v2 8x60K Microarray 039381 (Probe Name version) |
|
GSE165883
|
RNA-seq |
20 |
Illumina NextSeq 500 (Homo sapiens) |
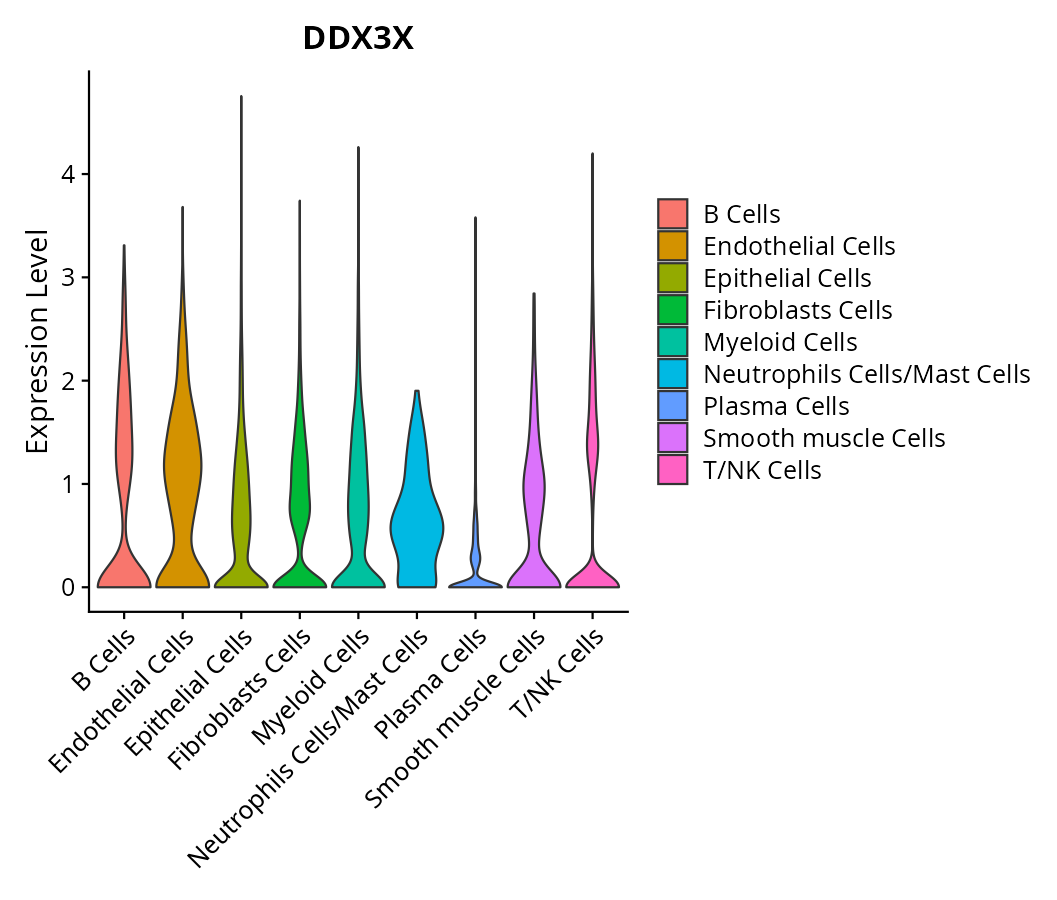
When the query gene is differentially changed in the dataset, a feature/violin plot will be displayed.
> Dataset: GSE197461 - Gene expression in cell subsets
|
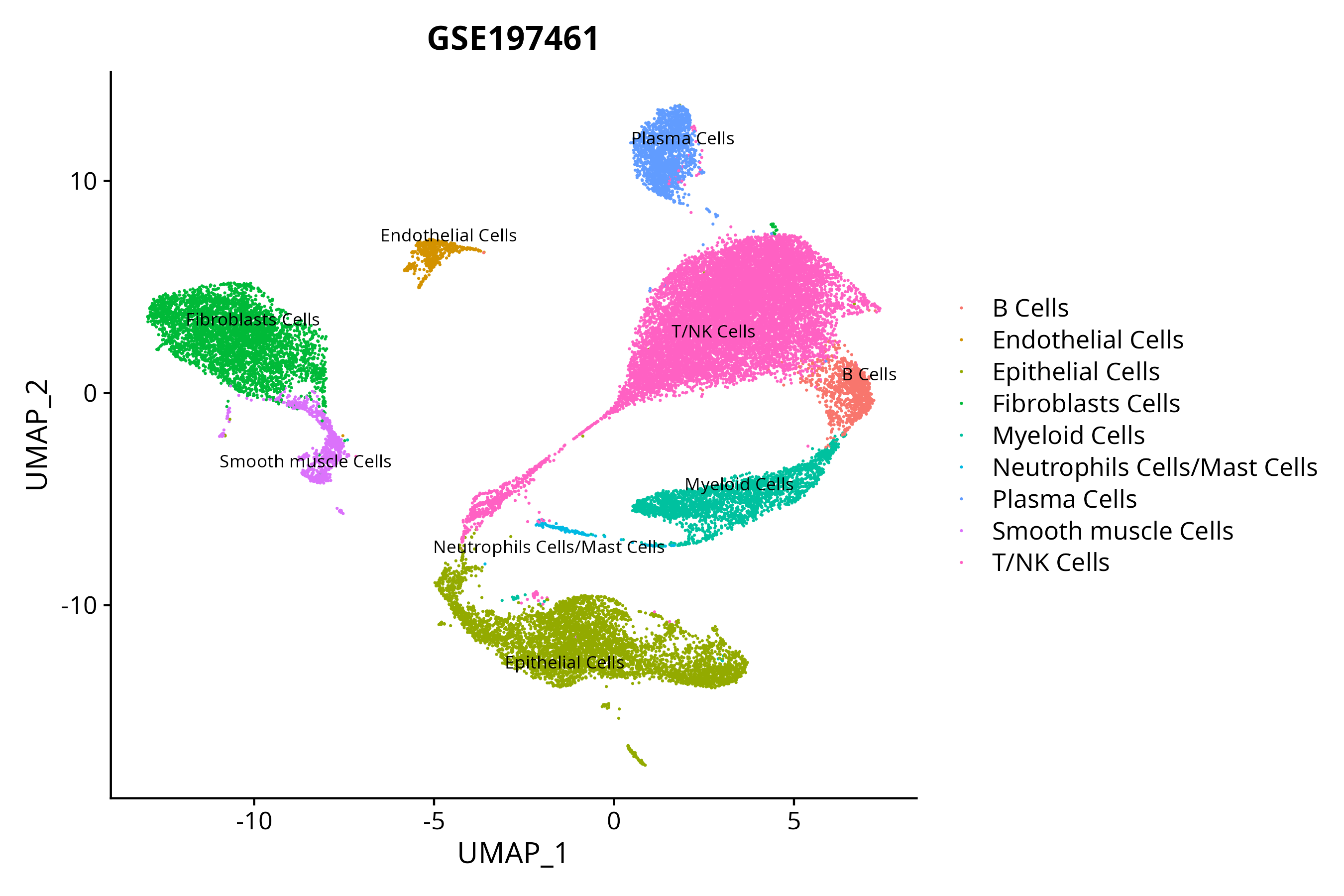
|
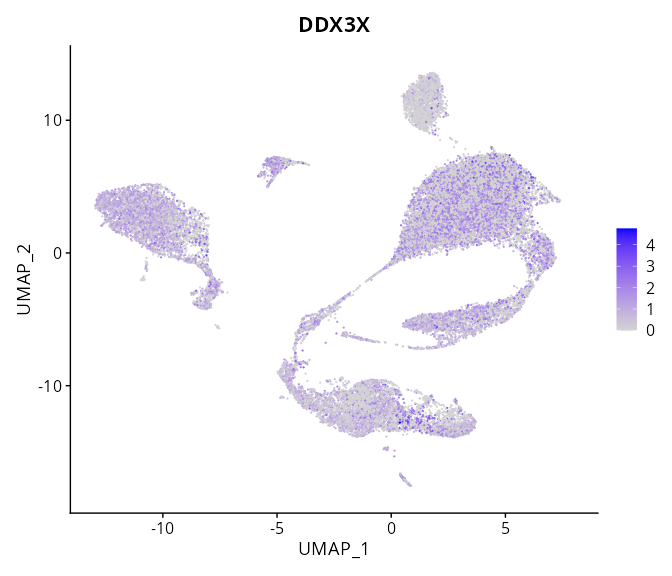
|
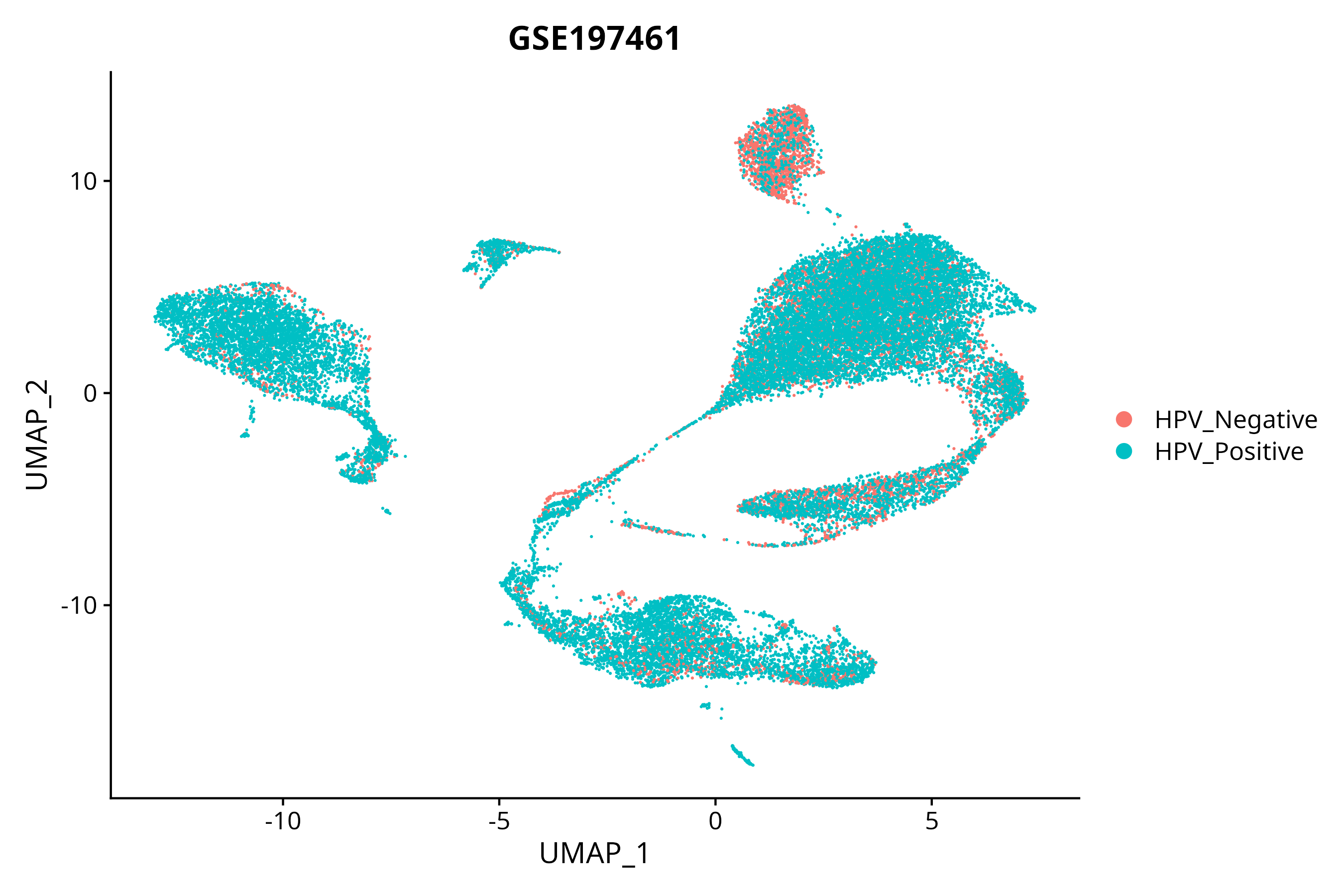
|
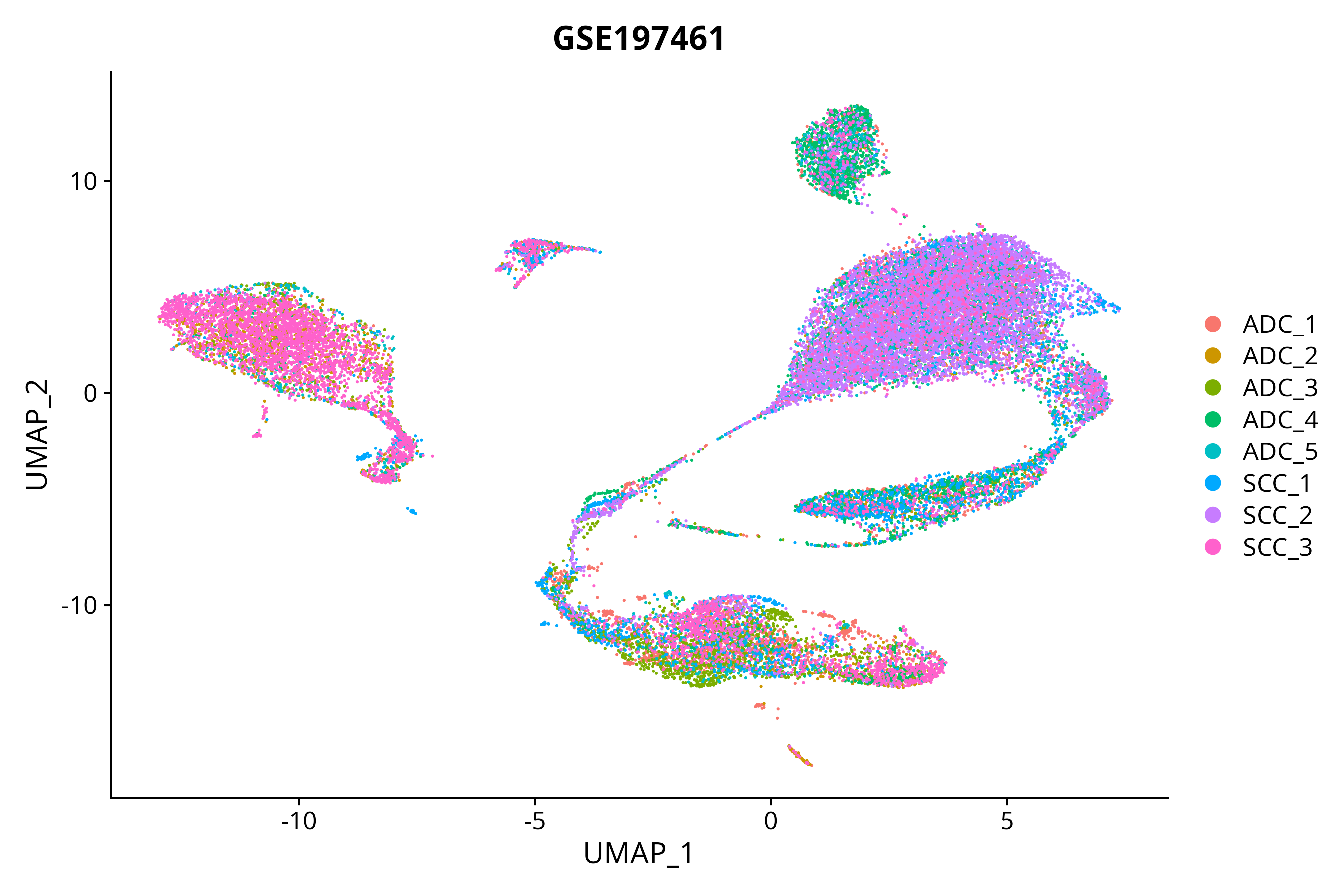
|
|
|
|
 HPV Target gene Detail Information
HPV Target gene Detail Information