Gene Information
|
Gene Name
|
EFNA1 |
|
Gene ID
|
1942
|
|
Gene Full Name
|
ephrin A1 |
|
Gene Alias
|
B61|ECKLG|EFL1|EPLG1|GMAN|LERK-1|LERK1|TNFAIP4 |
|
Transcripts
|
ENSG00000169242
|
|
Virus
|
HBV |
|
Gene Type
|
protein-coding |
|
HPA Location Info
|
Secreted to blood; 
|
|
Membrane Info
|
Cancer-related genes, Predicted secreted proteins, RAS pathway related proteins |
|
Uniport_ID
|
P20827
|
|
HGNC ID
|
HGNC:3221
|
|
OMIM ID
|
191164 |
|
Summary
|
This gene encodes a member of the ephrin (EPH) family. The ephrins and EPH-related receptors comprise the largest subfamily of receptor protein-tyrosine kinases and have been implicated in mediating developmental events, especially in the nervous system and in erythropoiesis. Based on their structures and sequence relationships, ephrins are divided into the ephrin-A (EFNA) class, which are anchored to the membrane by a glycosylphosphatidylinositol linkage, and the ephrin-B (EFNB) class, which are transmembrane proteins. This gene encodes an EFNA class ephrin which binds to the EPHA2, EPHA4, EPHA5, EPHA6, and EPHA7 receptors. Two transcript variants that encode different isoforms were identified through sequence analysis. [provided by RefSeq, Jul 2008] |
Target gene [EFNA1] related to VISs 
Integration Table: if previous studies reported that target gene was altered by virus integration events, the overlap between VISs in this literature and Cistrome factors was listed in this section
| DVID |
Chromosome |
HM |
TFBS |
CA |
Sum of Overlapped Records |
Detail |
| 1019982 |
chr1 |
39 |
0 |
0 |
39 |
View |
Target gene [EFNA1] related to Omics data
| Data ID |
Experiment type |
Sample number |
Platform |
|
GSE252863
|
scRNA-seq |
10 |
Illumina HiSeq 2500 (Homo sapiens) |
|
GSE236281
|
RNA-seq |
12 |
Illumina MiSeq (Homo sapiens) |
|
C GSE35465
|
Chip-seq;RNA-seq |
6 |
Illumina HiSeq 2000 (Homo sapiens) |
|
S GSE199850
|
scRNA-seq |
1 |
HiSeq X Ten (Homo sapiens) |
|
C GSE68402
|
Chip-seq |
26 |
Illumina MiSeq (Homo sapiens);Illumina HiSeq 2500 (Homo sapiens) |
|
TCGA_LIHC_HBV
|
DNA methylation sequencing;RNA-seq |
97 |
TCGA |
|
C GSE270130
|
Chip-seq |
27 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE224901
|
RNA-seq |
21 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE100400
|
Chip-seq;RNA-seq;4C_cccDNA |
31 |
Illumina NextSeq 500 (Homo sapiens);Illumina NextSeq 500 (Mus musculus) |
|
GSE173897
|
RNA-seq |
95 |
Illumina HiSeq 4000 (Homo sapiens) |
|
GSE262515
|
RNA-seq |
21 |
Illumina HiSeq 2500 (Homo sapiens);Illumina HiSeq 2500 (Mus musculus) |
|
GSE110345
|
RNA-seq |
4 |
Illumina HiSeq 2500 (Homo sapiens) |
|
C GSE131257
|
ATAC-seq;RNA-seq |
19 |
Illumina HiSeq 2500 (Homo sapiens) |
|
GSE94660
|
RNA-seq |
42 |
Illumina HiSeq 2500 (Homo sapiens) |
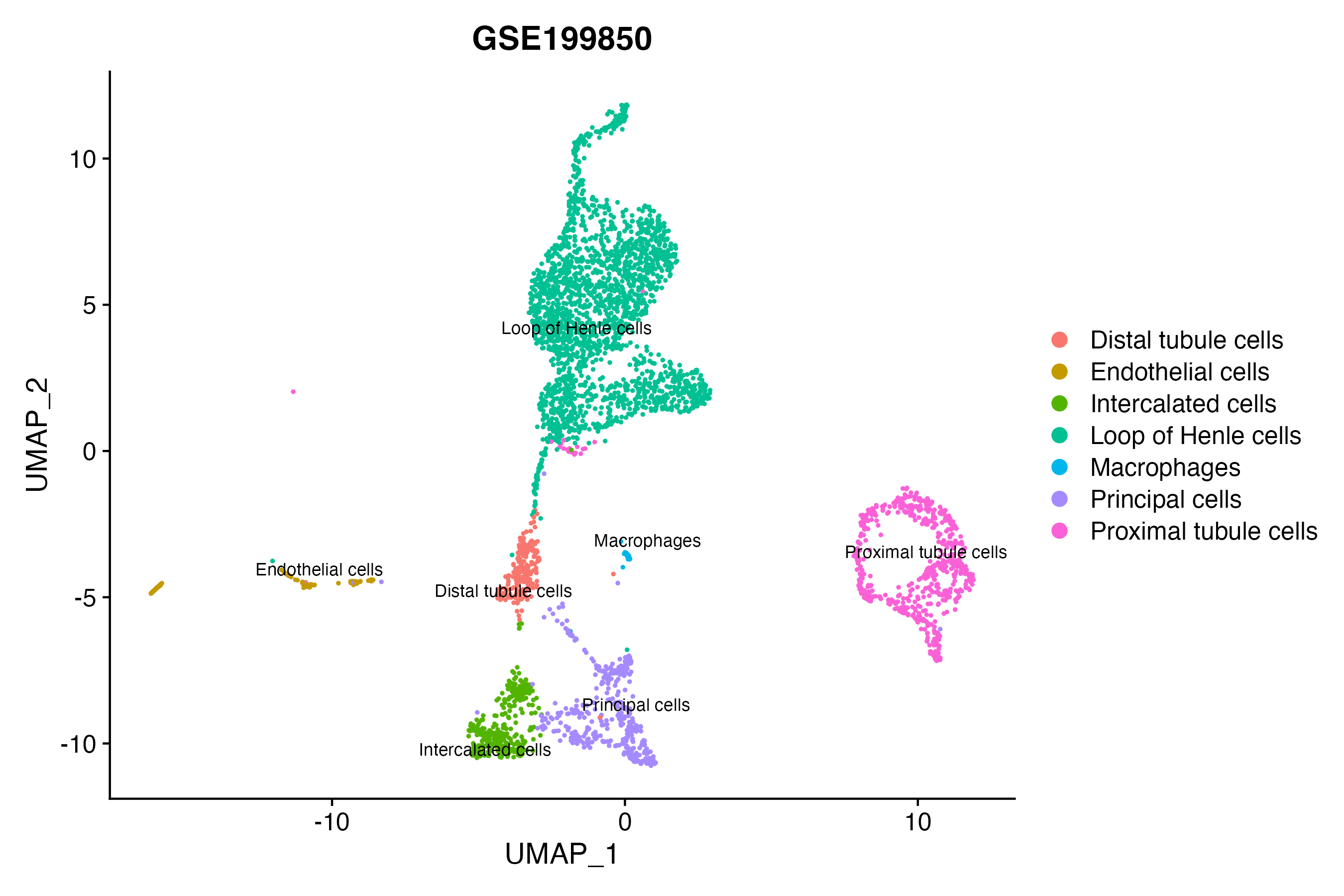
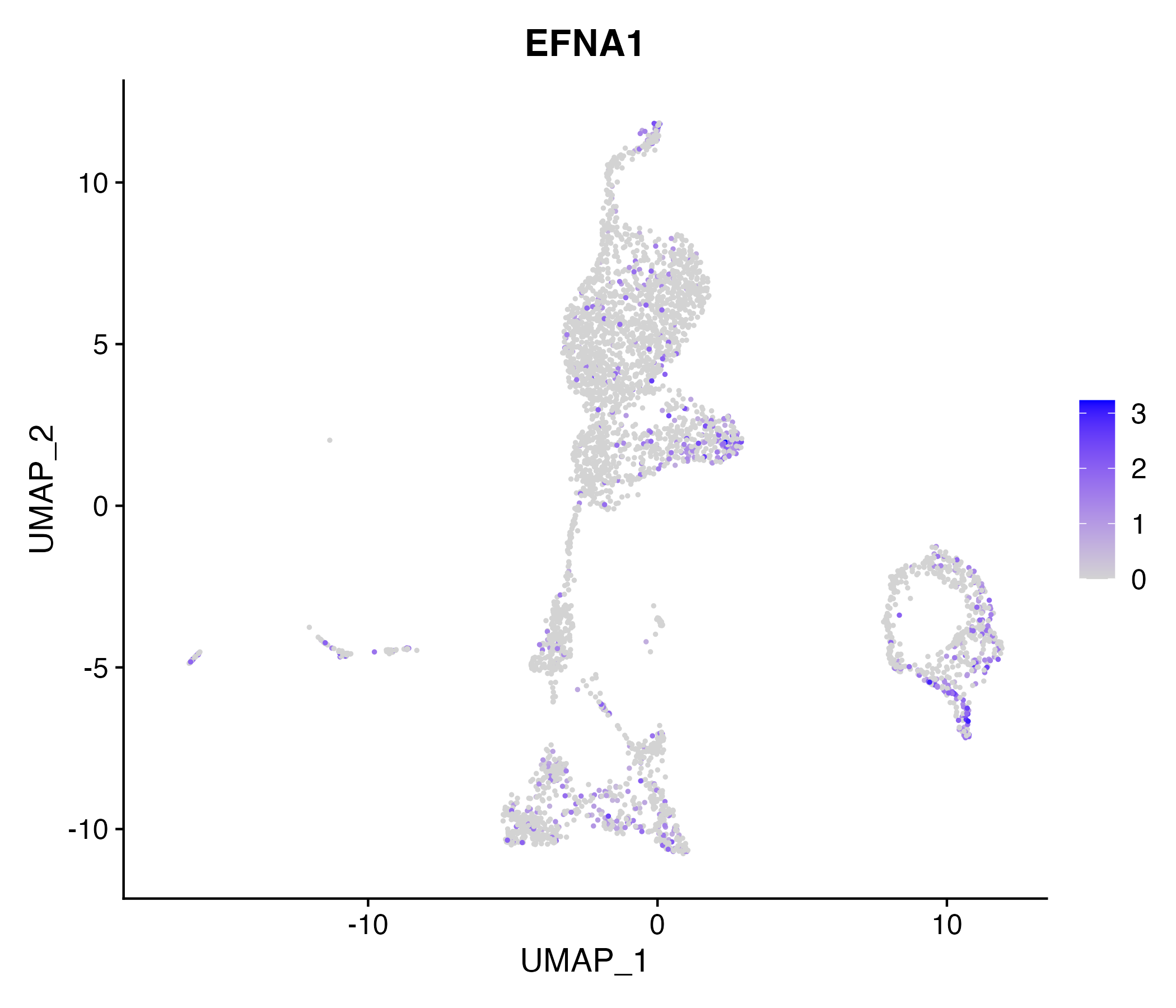
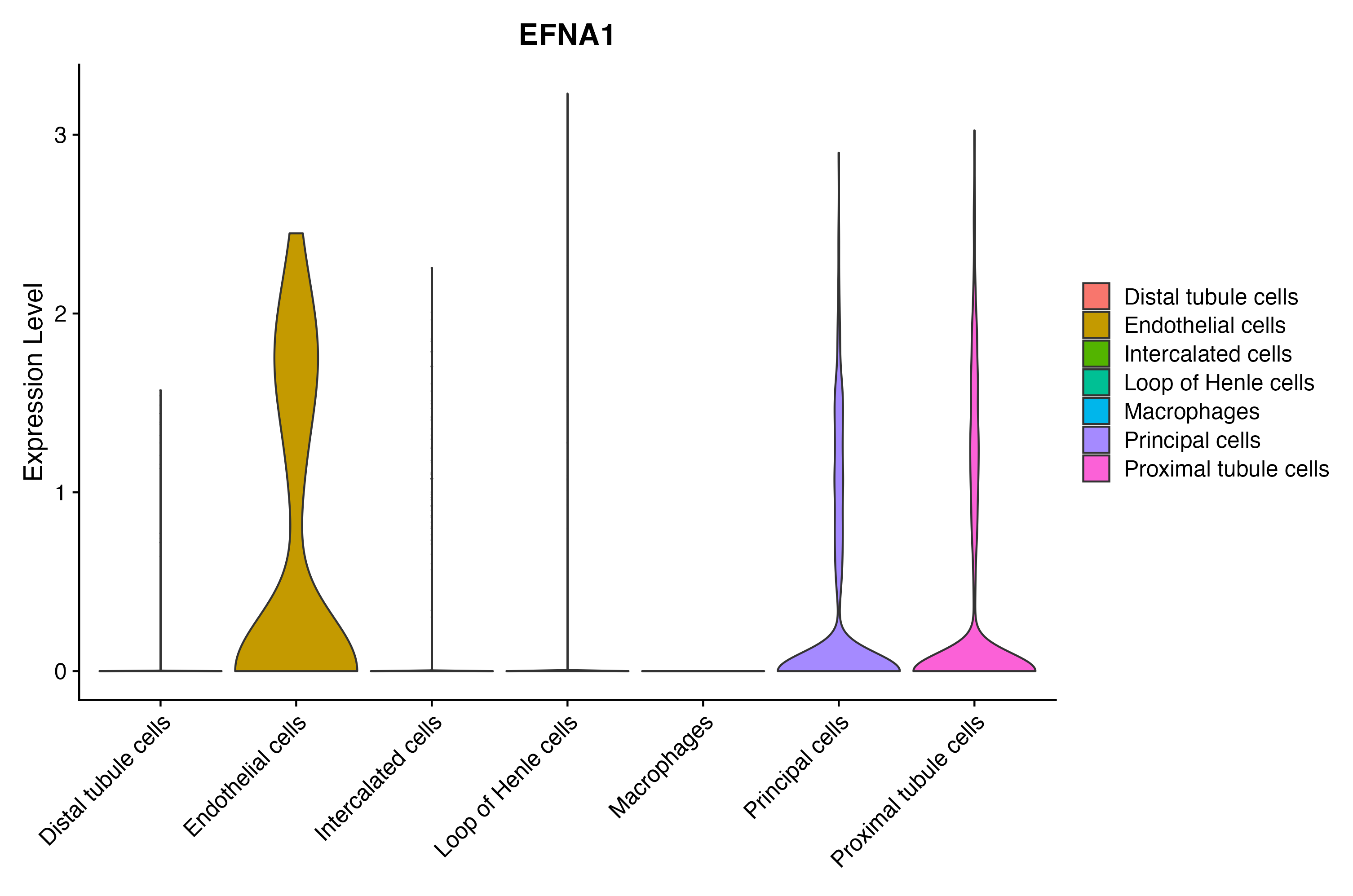
When the query gene is differentially changed in the dataset, a feature/violin plot will be displayed.
> Dataset: GSE199850 - Gene expression in cell subsets
|
|
|
|
|
|
|
|
 HBV Target gene Detail Information
HBV Target gene Detail Information