Gene Information
|
Gene Name
|
KCNAB1 |
|
Gene ID
|
7881
|
|
Gene Full Name
|
potassium voltage-gated channel subfamily A regulatory beta subunit 1 |
|
Gene Alias
|
AKR6A3|KCNA1B|KV-BETA-1|Kvb1.3|hKvBeta3|hKvb3 |
|
Transcripts
|
ENSG00000169282
|
|
Virus
|
HBV |
|
Gene Type
|
protein-coding |
|
HPA Location Info
|
Vesicles;Nucleoplasm, Plasma membrane; 
|
|
Membrane Info
|
Predicted intracellular proteins, Transporters |
|
Uniport_ID
|
Q14722
|
|
HGNC ID
|
HGNC:6228
|
|
OMIM ID
|
601141 |
|
Summary
|
Potassium channels represent the most complex class of voltage-gated ion channels from both functional and structural standpoints. Their diverse functions include regulating neurotransmitter release, heart rate, insulin secretion, neuronal excitability, epithelial electrolyte transport, smooth muscle contraction, and cell volume. Four sequence-related potassium channel genes - shaker, shaw, shab, and shal - have been identified in Drosophila, and each has been shown to have human homolog(s). This gene encodes a member of the potassium channel, voltage-gated, shaker-related subfamily. This member includes distinct isoforms which are encoded by alternatively spliced transcript variants of this gene. Some of these isoforms are beta subunits, which form heteromultimeric complexes with alpha subunits and modulate the activity of the pore-forming alpha subunits. [provided by RefSeq, Apr 2015] |
Target gene [KCNAB1] related to VISs 
Integration Table: if previous studies reported that target gene was altered by virus integration events, the overlap between VISs in this literature and Cistrome factors was listed in this section
| DVID |
Chromosome |
HM |
TFBS |
CA |
Sum of Overlapped Records |
Detail |
| 1002738 |
chr3 |
36 |
36 |
140 |
212 |
View |
| 1006960 |
chr3 |
3 |
14 |
15 |
32 |
View |
| 1024665 |
chr3 |
17 |
1 |
1 |
19 |
View |
| 1041368 |
chr3 |
16 |
9 |
0 |
25 |
View |
Target gene [KCNAB1] related to Omics data
| Data ID |
Experiment type |
Sample number |
Platform |
|
GSE236281
|
RNA-seq |
12 |
Illumina MiSeq (Homo sapiens) |
|
C GSE68402
|
Chip-seq |
26 |
Illumina MiSeq (Homo sapiens);Illumina HiSeq 2500 (Homo sapiens) |
|
TCGA_LIHC_HBV
|
DNA methylation sequencing;RNA-seq |
97 |
TCGA |
|
GSE224901
|
RNA-seq |
21 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
C GSE100400
|
Chip-seq;RNA-seq;4C_cccDNA |
31 |
Illumina NextSeq 500 (Homo sapiens);Illumina NextSeq 500 (Mus musculus) |
|
GSE173897
|
RNA-seq |
95 |
Illumina HiSeq 4000 (Homo sapiens) |
|
GSE262515
|
RNA-seq |
21 |
Illumina HiSeq 2500 (Homo sapiens);Illumina HiSeq 2500 (Mus musculus) |
|
GSE110345
|
RNA-seq |
4 |
Illumina HiSeq 2500 (Homo sapiens) |
|
C GSE131257
|
ATAC-seq;RNA-seq |
19 |
Illumina HiSeq 2500 (Homo sapiens) |
|
E GSE94660
|
RNA-seq |
42 |
Illumina HiSeq 2500 (Homo sapiens) |
When the query gene is differentially changed in the dataset, a volcano/bar plot will be displayed.
> Dataset: GSE94660 - KCNAB1 expression across samples
|
Volcano Plot
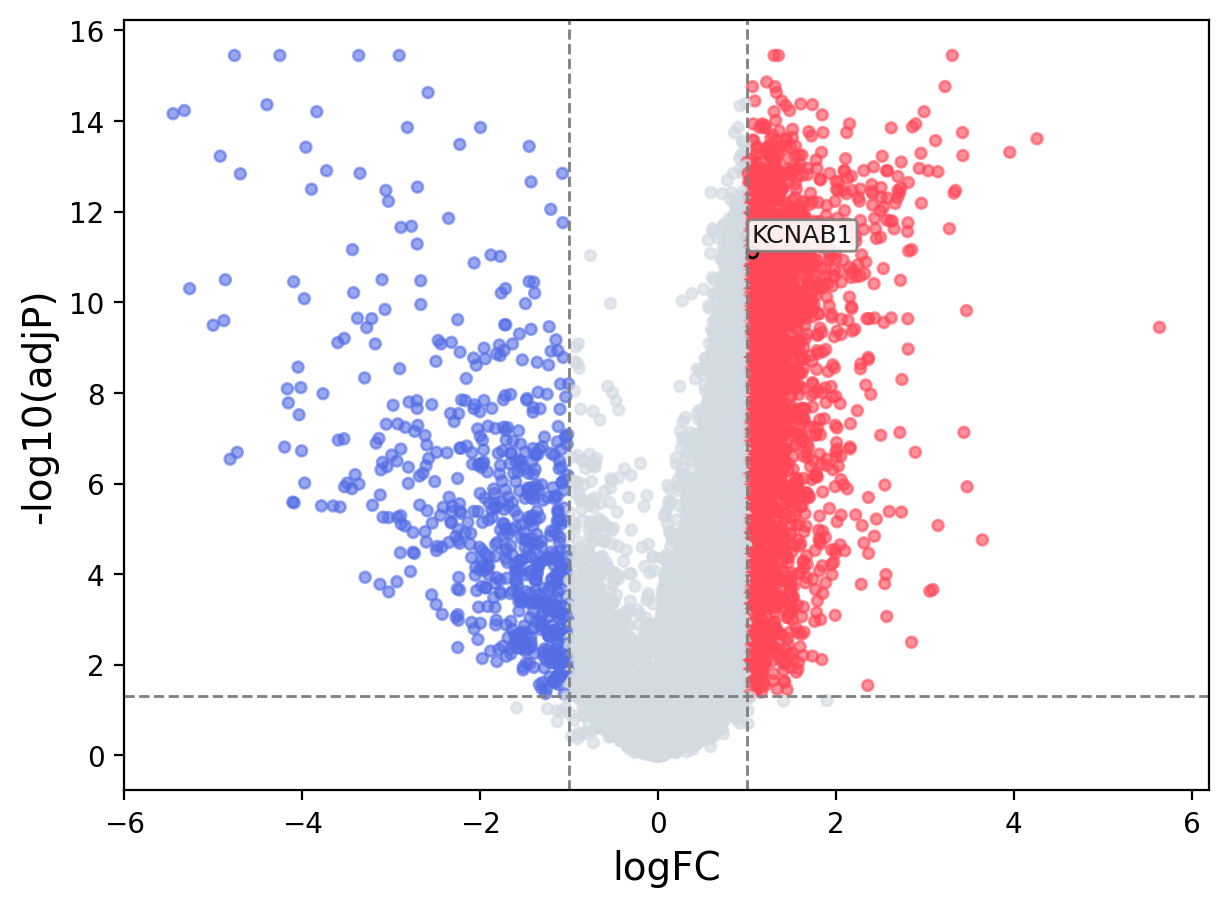
|
Bar Plot
|
|
|
 HBV Target gene Detail Information
HBV Target gene Detail Information