Gene Information
|
Gene Name
|
KCNN2 |
|
Gene ID
|
3781
|
|
Gene Full Name
|
potassium calcium-activated channel subfamily N member 2 |
|
Gene Alias
|
DYT34|KCa2.2|NEDMAB|SK2|SKCA2|SKCa 2|hSK2 |
|
Transcripts
|
ENSG00000080709
|
|
Virus
|
HBV |
|
Gene Type
|
protein-coding |
|
HPA Location Info
|
Nucleoplasm, Plasma membrane; 
|
|
Membrane Info
|
Disease related genes, FDA approved drug targets, Human disease related genes, Plasma proteins, Predicted membrane proteins, Transporters, Voltage-gated ion channels |
|
Uniport_ID
|
Q9H2S1
|
|
HGNC ID
|
HGNC:6291
|
|
OMIM ID
|
605879 |
|
Summary
|
Action potentials in vertebrate neurons are followed by an afterhyperpolarization (AHP) that may persist for several seconds and may have profound consequences for the firing pattern of the neuron. Each component of the AHP is kinetically distinct and is mediated by different calcium-activated potassium channels. The protein encoded by this gene is activated before membrane hyperpolarization and is thought to regulate neuronal excitability by contributing to the slow component of synaptic AHP. This gene is a member of the KCNN family of potassium channel genes. The encoded protein is an integral membrane protein that forms a voltage-independent calcium-activated channel with three other calmodulin-binding subunits. Alternate splicing of this gene results in multiple transcript variants. [provided by RefSeq, May 2013] |
Target gene [KCNN2] related to VISs 
Integration Table: if previous studies reported that target gene was altered by virus integration events, the overlap between VISs in this literature and Cistrome factors was listed in this section
| DVID |
Chromosome |
HM |
TFBS |
CA |
Sum of Overlapped Records |
Detail |
| 1013363 |
chr5 |
3 |
2 |
0 |
5 |
View |
| 1014805 |
chr5 |
0 |
0 |
3 |
3 |
View |
| 1016787 |
chr5 |
0 |
0 |
1 |
1 |
View |
| 1018905 |
chr5 |
0 |
0 |
0 |
0 |
View |
Target gene [KCNN2] related to Omics data
| Data ID |
Experiment type |
Sample number |
Platform |
|
GSE236281
|
RNA-seq |
12 |
Illumina MiSeq (Homo sapiens) |
|
C GSE35465
|
Chip-seq;RNA-seq |
6 |
Illumina HiSeq 2000 (Homo sapiens) |
|
C GSE68402
|
Chip-seq |
26 |
Illumina MiSeq (Homo sapiens);Illumina HiSeq 2500 (Homo sapiens) |
|
TCGA_LIHC_HBV
|
DNA methylation sequencing;RNA-seq |
97 |
TCGA |
|
C GSE270130
|
Chip-seq |
27 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE224901
|
RNA-seq |
21 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
C GSE100400
|
Chip-seq;RNA-seq;4C_cccDNA |
31 |
Illumina NextSeq 500 (Homo sapiens);Illumina NextSeq 500 (Mus musculus) |
|
GSE173897
|
RNA-seq |
95 |
Illumina HiSeq 4000 (Homo sapiens) |
|
GSE262515
|
RNA-seq |
21 |
Illumina HiSeq 2500 (Homo sapiens);Illumina HiSeq 2500 (Mus musculus) |
|
GSE110345
|
RNA-seq |
4 |
Illumina HiSeq 2500 (Homo sapiens) |
|
C GSE131257
|
ATAC-seq;RNA-seq |
19 |
Illumina HiSeq 2500 (Homo sapiens) |
|
E GSE94660
|
RNA-seq |
42 |
Illumina HiSeq 2500 (Homo sapiens) |
When the query gene is differentially changed in the dataset, a volcano/bar plot will be displayed.
> Dataset: GSE94660 - KCNN2 expression across samples
|
Volcano Plot
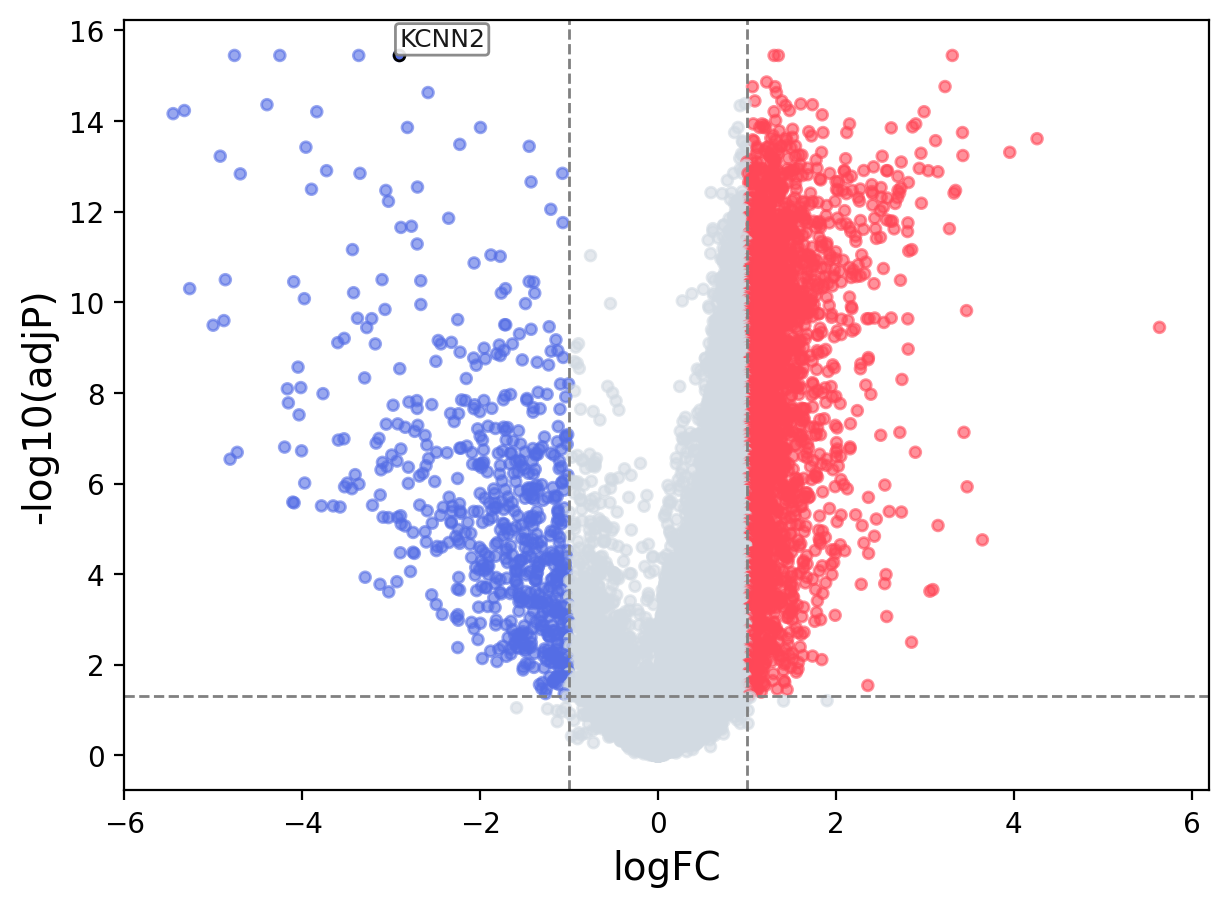
|
Bar Plot
|
|
|
 HBV Target gene Detail Information
HBV Target gene Detail Information