Gene Information
|
Gene Name
|
POR |
|
Gene ID
|
5447
|
|
Gene Full Name
|
cytochrome p450 oxidoreductase |
|
Gene Alias
|
CPR|CYPOR|P450R |
|
Transcripts
|
ENSG00000127948
|
|
Virus
|
HBV |
|
Gene Type
|
protein-coding |
|
HPA Location Info
|
Vesicles;Nucleoplasm, Cytosol; 
|
|
Membrane Info
|
Disease related genes, Enzymes, Human disease related genes, Metabolic proteins, Plasma proteins, Potential drug targets, Predicted intracellular proteins, Predicted membrane proteins |
|
Uniport_ID
|
P16435
|
|
HGNC ID
|
HGNC:9208
|
|
OMIM ID
|
124015 |
|
Summary
|
This gene encodes an endoplasmic reticulum membrane oxidoreductase that is essential for multiple metabolic processes, including reactions catalyzed by cytochrome P450 proteins for metabolism of steroid hormones, drugs and xenobiotics. The encoded protein has a flavin adenine dinucleotide (FAD)-binding domain and a flavodoxin-like domain which bind two cofactors, FAD and FMN, that allow it to donate electrons directly from NADPH to all microsomal P450 enzymes. Mutations in this gene cause a complex set of disorders, including apparent combined P450C17 and P450C21 deficiency, amenorrhea and disordered steroidogenesis, congenital adrenal hyperplasia and Antley-Bixler syndrome, that resemble those caused by defects in steroid metabolizing enzymes such as aromatase, 21-hydroxylase, and 17 alpha-hydroxylase. [provided by RefSeq, Aug 2020] |
Target gene [POR] related to VISs 
Integration Table: if previous studies reported that target gene was altered by virus integration events, the overlap between VISs in this literature and Cistrome factors was listed in this section
Target gene [POR] related to Omics data
| Data ID |
Experiment type |
Sample number |
Platform |
|
GSE236281
|
RNA-seq |
12 |
Illumina MiSeq (Homo sapiens) |
|
C GSE35465
|
Chip-seq;RNA-seq |
6 |
Illumina HiSeq 2000 (Homo sapiens) |
|
GSE199850
|
scRNA-seq |
1 |
HiSeq X Ten (Homo sapiens) |
|
C GSE68402
|
Chip-seq |
26 |
Illumina MiSeq (Homo sapiens);Illumina HiSeq 2500 (Homo sapiens) |
|
GSE247322
|
scRNA-seq |
27 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
TCGA_LIHC_HBV
|
DNA methylation sequencing;RNA-seq |
97 |
TCGA |
|
C GSE270130
|
Chip-seq |
27 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE224901
|
RNA-seq |
21 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE100400
|
Chip-seq;RNA-seq;4C_cccDNA |
31 |
Illumina NextSeq 500 (Homo sapiens);Illumina NextSeq 500 (Mus musculus) |
|
GSE173897
|
RNA-seq |
95 |
Illumina HiSeq 4000 (Homo sapiens) |
|
GSE262515
|
RNA-seq |
21 |
Illumina HiSeq 2500 (Homo sapiens);Illumina HiSeq 2500 (Mus musculus) |
|
GSE110345
|
RNA-seq |
4 |
Illumina HiSeq 2500 (Homo sapiens) |
|
C GSE131257
|
ATAC-seq;RNA-seq |
19 |
Illumina HiSeq 2500 (Homo sapiens) |
|
GSE94660
|
RNA-seq |
42 |
Illumina HiSeq 2500 (Homo sapiens) |
When the gene can detect a peak in the dataset, a peak plot will be displayed.
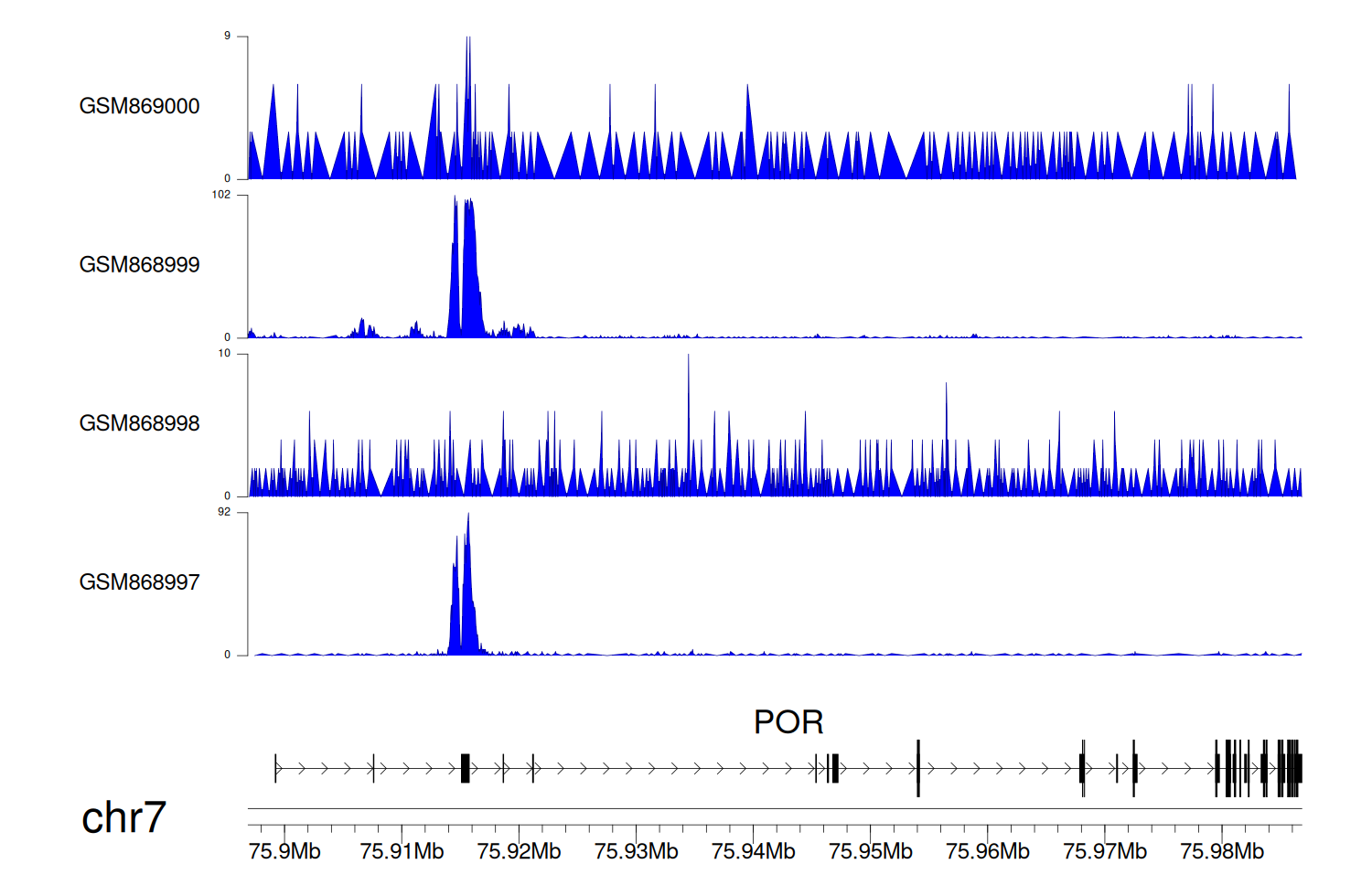
> Dataset: GSE35465 - POR peak across samples
|
Peak Plot
|
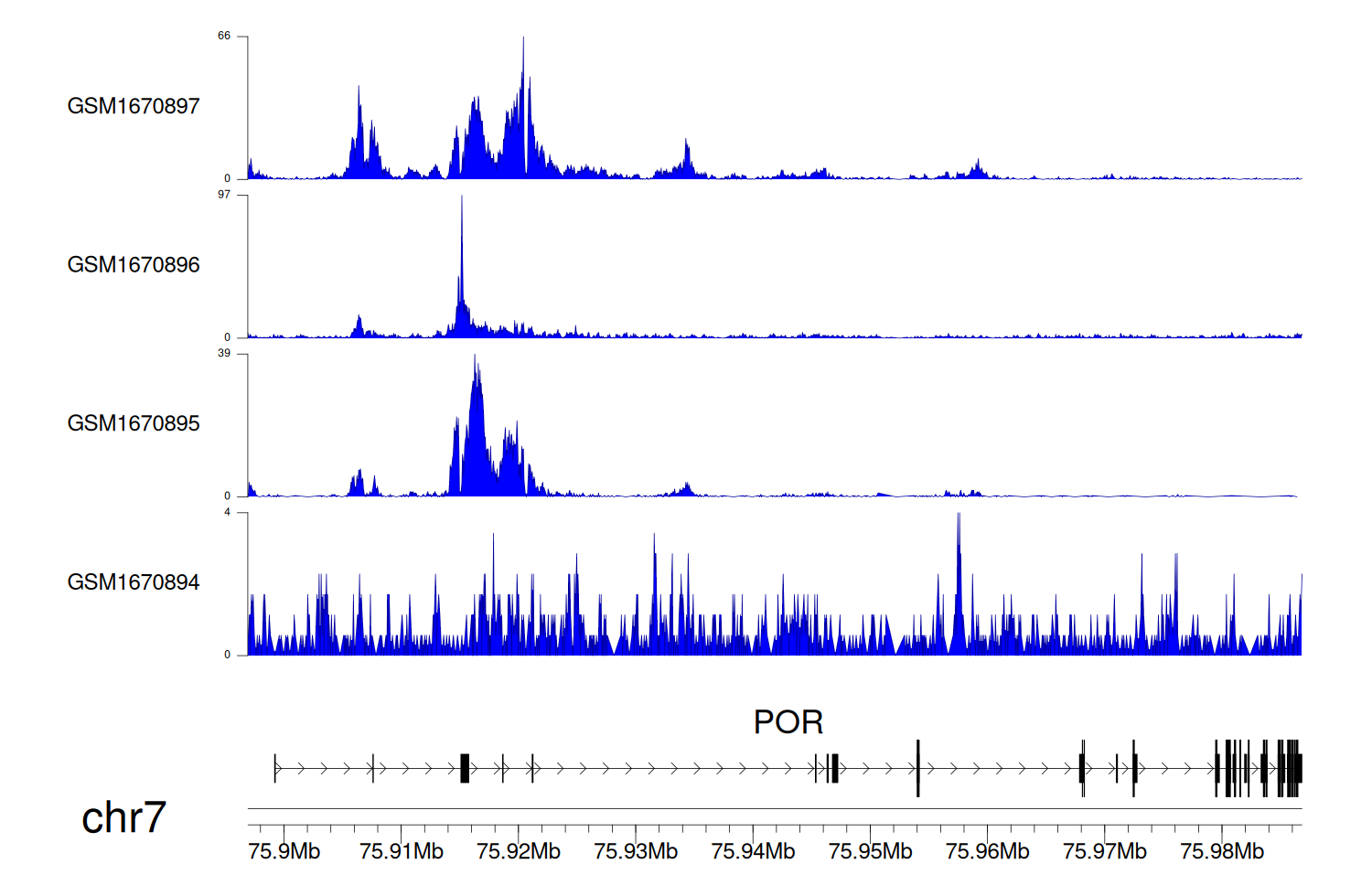
> Dataset: GSE68402 - POR peak across samples
|
Peak Plot
|
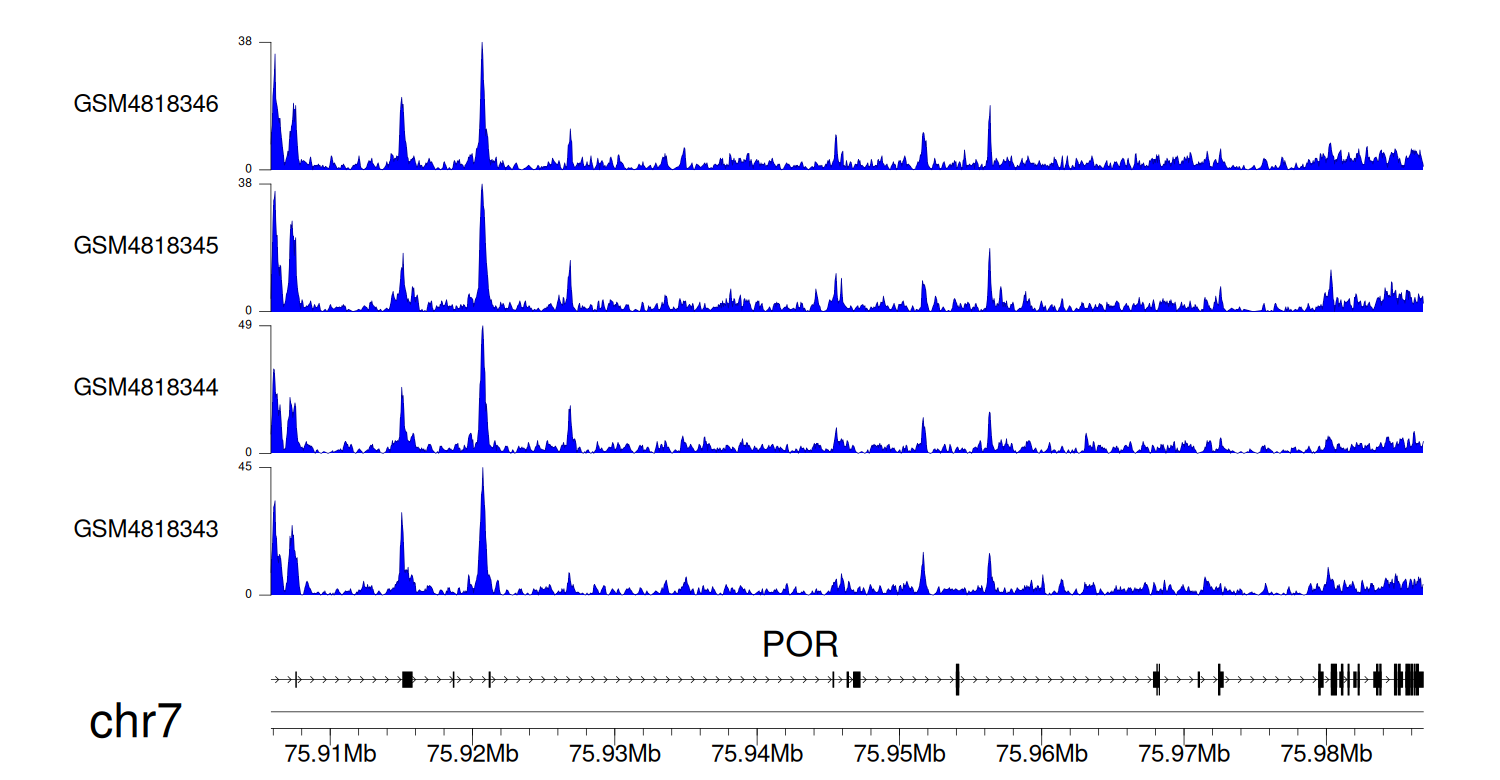
> Dataset: GSE270130 - POR peak across samples
|
Peak Plot
|
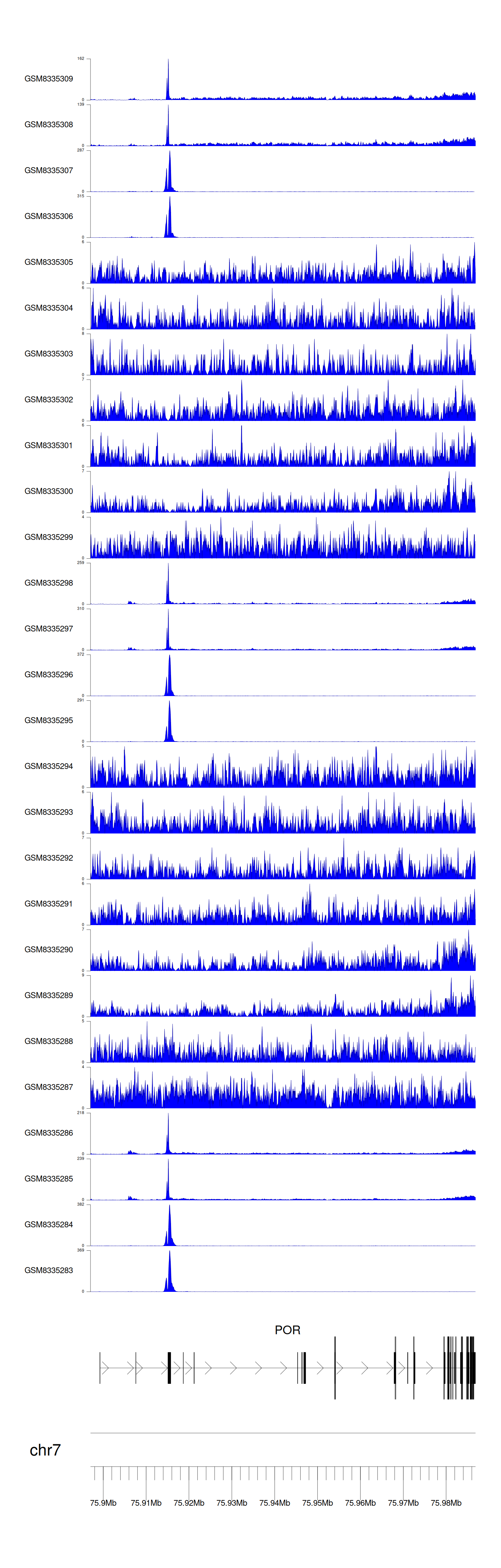
> Dataset: GSE131257 - POR peak across samples
|
Peak Plot
|
|
|
 HBV Target gene Detail Information
HBV Target gene Detail Information